Food Safety & Drug Residues Kits
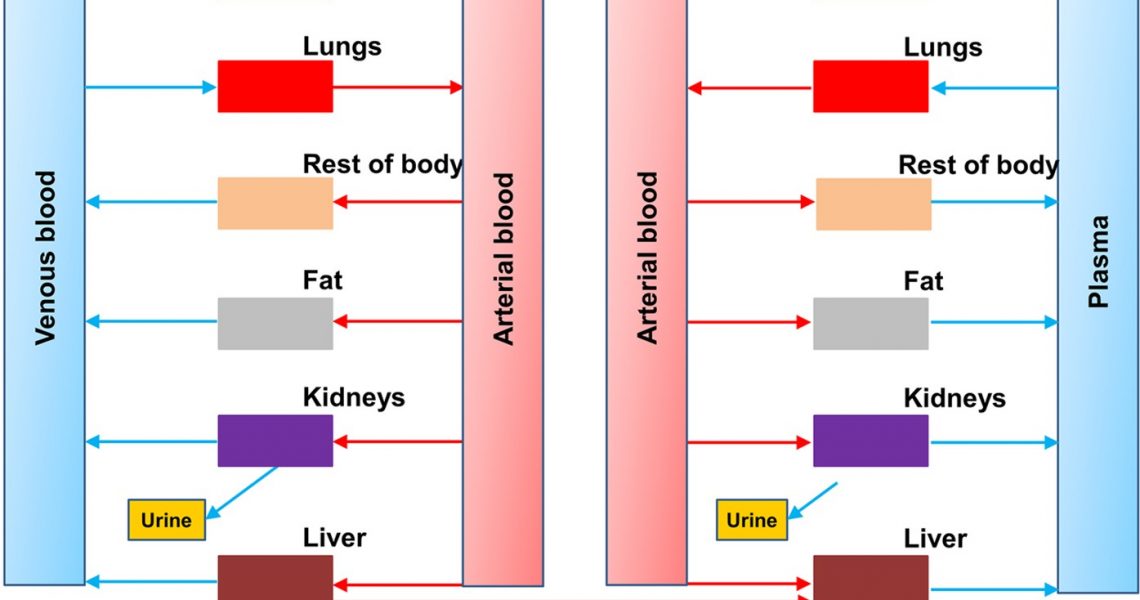
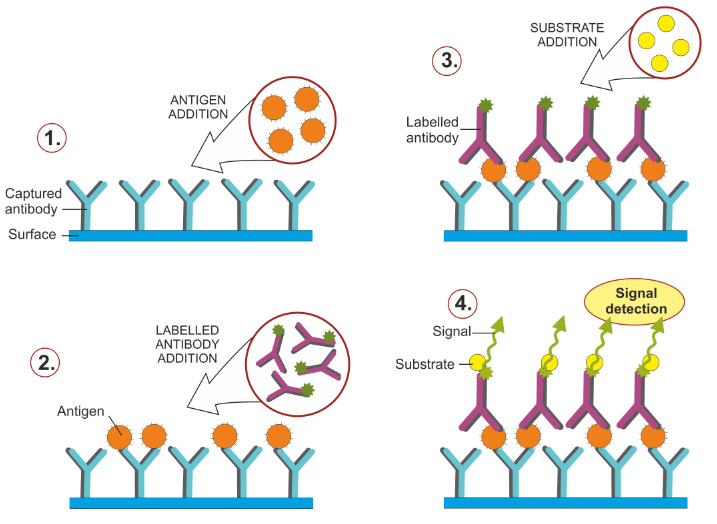
Drug Residues and Microbial Contamination in Food: Surveillance and Compliance A primary goal of US food safety programs is the control of contaminants that may appear in food due to drug use in animals or the inadvertent introduction of microorganisms. Drug residue control and surveillance for microbial contamination are achieved through an extensive and rigorous process of sampling, testing, reporting and compliance. Tens of thousands […]